The complement system apparently engulfs a network of proteins that enhance the work of antibodies in destroying foreign invaders. It probably acts as a rapid defense mechanism in innumerable species from primitive sponges to humans. A team of experts from the University of Pennsylvania School of Medicine and Utrecht University has detailed the atomic structure of two key transient enzyme complexes in the human complement system.
Complement proteins seem to point out bacterial and dying host cells for elimination by the body’s cellular cleanup services. These protein have been supposedly implicated in 30 diseases, including stroke, myocardial infarction, and age-related macular degeneration. The research findings can therefore aid in designing novel drug therapeutics. The complement system appears as a form of ‘innate’ or generic immunity that demonstrates a complex biochemical network of molecular and cellular communication events. Such events may be responsible for the destruction and elimination of pathogens as well as damaged cells.
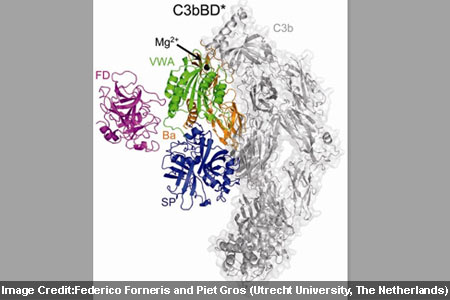
At the time of the research, investigators analyzed two enzyme complexes known as C3bB and C3bBD that drive a central step in amplifying the response by complement proteins. The complement protein C3 presumably is enzymatically cleaved to form C3b for binding with the surface of a target cell. Once formed into C3b, it is believed to bind with factor B for producing C3bB. This complex possibly binds another enzyme, factor D and produces C3bBD. It appears that the major target of the active C3bBb is C3 itself that leads to rapid amplification of the complement cascade.
John Lambris, PhD, added, “Besides shedding light on a highly elegant mechanism of concerted activation and intrinsic regulation, this work also offers a detailed insight into one of the most important therapeutic targets within the complement network, which may facilitate rational drug development and could lead to novel drugs for treating complement-related diseases.”
In order to capture structural snapshots of C3bB and C3bBD, experts generated mutant proteins that would stabilize the complexes in their active forms. Then by means of x-ray crystallography the two complexes in atomic were described in detail. On binding to C3b, factor B supposedly modifies its shape to form an ‘open complex’ that can be bound by factor D. The factor D may now shift its shape in a more subtle yet crucial way. The free protein seems to be inactive as a protein loop blocks the active site of the enzyme. After binding to C3bB, that loop apparently changes its position and activates factor D to split C3bB into the C3bBb complex. The findings presumably provide information for developing inhibitors against factor D for treating complement-associated diseases.
The research was published in the December 24 issue of Science.
